Section 6 RCBD Example: SAS
6.1 Load and Explore Data
These data are from a winter wheat variety trial in Alliance, Nebraska (Stroup, Baenziger, and Mulitze 1994) and are commonly used to demonstrate spatial adjustment in linear modeling. The data represent yields of 56 varieties originally laid out in a randomized complete block design (RCB). The local topography combined with winter kill, however, induced spatial variability that did not correspond to the RCB design (Stroup 2013) which produced biased estimates in a standard unadjusted analysis.
The code below reads the data from a CSV file located the GitHub repo for this book. The data file can be downloaded from here. In this file, missing values for yield are denoted as ‘NA’. These values are not actually missing, but represent blank plots separating blocks in the original design of the study. The PROC FORMAT section converts these to numeric missing values in SAS (defined as a single period). They are then removed from the data before proceeding to analyses. Also note that row and column indices are multiplied by constants to convert them to the meter dimensions of the plots. .
The first six observations are displayed. The data contains variables for variety (gen), replication (rep), yield, and column and row identifiers.
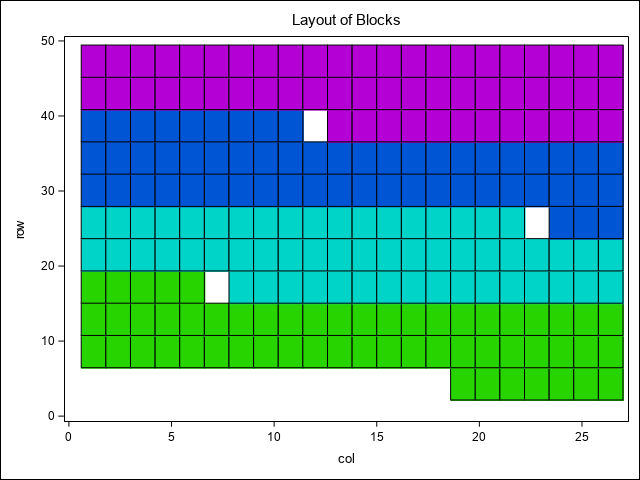
Last, a map of the 4 replications (blocks) from the original experimental design is shown.
proc format;
invalue has_NA
'NA' = .;
;
filename NIN url "https://raw.githubusercontent.com/IdahoAgStats/guide-to-field-trial-spatial-analysis/master/data/stroup_nin_wheat.csv";
data alliance;
infile NIN firstobs=2 delimiter=',';
informat yield has_NA.;
input entry $ rep $ yield col row;
Row = 4.3*Row;
Col = 1.2*Col;
if yield=. then delete;
run;
proc print data=alliance(obs=6);
title1 ' Alliance Nebraska Wheat Variety Data';
run;
ODS html GPATH = ".\img\" ;
proc sgplot data=alliance;
styleattrs datacolors=(cx28D400 cx00D4C8 cx0055D4 cxB400D4) datalinepatterns=(solid dash);
HEATMAPPARM y=row x=col COLORgroup=rep/ outline;
title1 'Layout of Blocks';
run;| Alliance Nebraska Wheat Variety Data |
| Obs | yield | entry | rep | col | row |
|---|---|---|---|---|---|
| 1 | 29.25 | Lancer | R1 | 19.2 | 4.3 |
| 2 | 31.55 | Brule | R1 | 20.4 | 4.3 |
| 3 | 35.05 | Redland | R1 | 21.6 | 4.3 |
| 4 | 30.10 | Cody | R1 | 22.8 | 4.3 |
| 5 | 33.05 | Arapahoe | R1 | 24.0 | 4.3 |
| 6 | 30.25 | NE83404 | R1 | 25.2 | 4.3 |
6.1.1 Plots of Field Trends
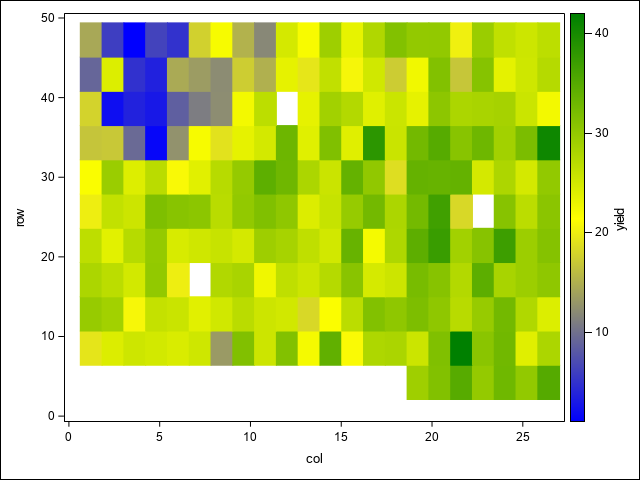
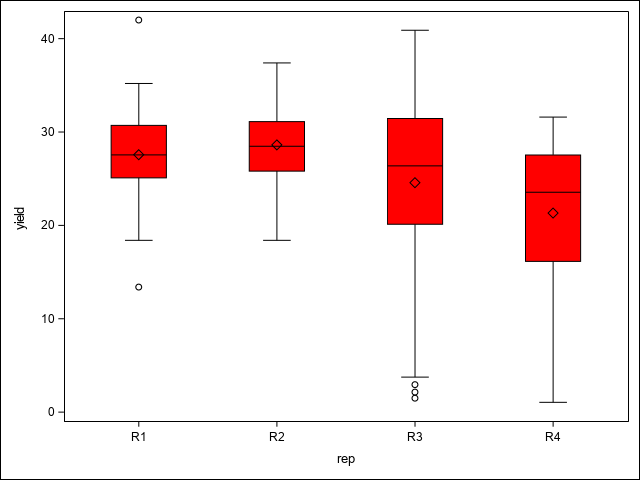
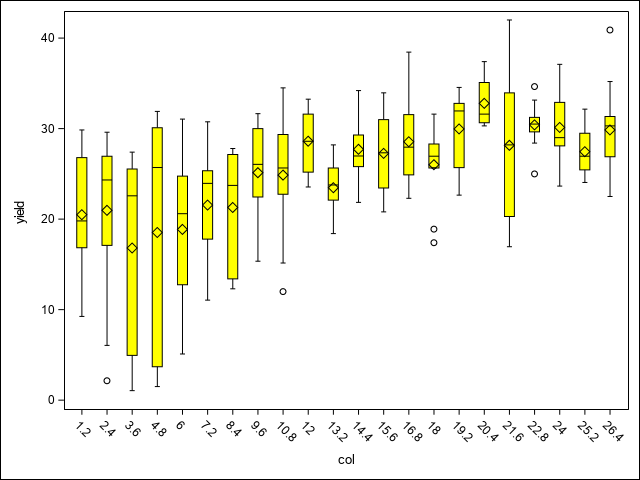
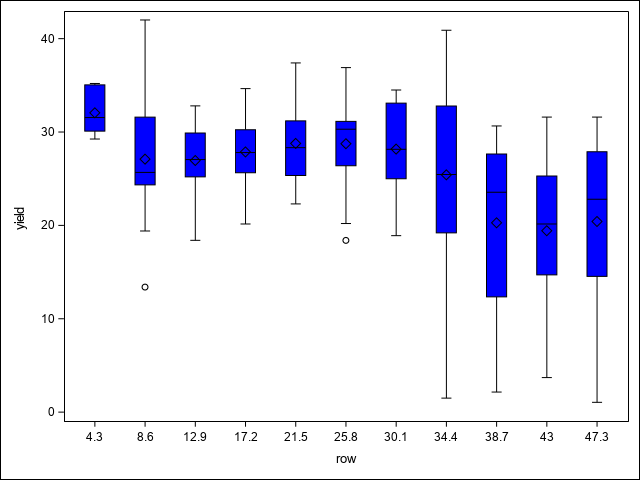
A first step in assessing spatial variability is to plot the data to visually assess any trends or patterns. This code uses the SGPLOT procedure to examine spatial patterns through a heat map of yields, as well as potential trends across replications, columns, and rows using box plots.
ODS html GPATH = ".\img\" ;
proc sgplot data=alliance;
HEATMAPPARM y=Row x=Col COLORRESPONSE=yield/ colormodel=(blue yellow green);
run;
proc sgplot data=alliance;
vbox yield/category=rep FILLATTRS=(color=red) LINEATTRS=(color=black) WHISKERATTRS=(color=black);
run;
proc sgplot data=alliance;
vbox yield/category=Col FILLATTRS=(color=yellow) LINEATTRS=(color=black) WHISKERATTRS=(color=black);
run;
proc sgplot data=alliance;
vbox yield/category=Row FILLATTRS=(color=blue) LINEATTRS=(color=black) WHISKERATTRS=(color=black);
run;In the heat map, there is a notable region where yield dips in the north west corner of the study area. This area runs across the two top most blocks and is positioned towards their ends, making those blocks non-homogeneous. As Stroup (Stroup 2013) notes, this is due to a hilly area with low snow cover and high exposure to low winter temperatures. The box plots demonstrate this pattern across blocks, columns, and rows as well. From these initial graphics, it is clear there are discernible spatial patterns and trends present.
6.2 Estimating and Testing Spatial Correlation
6.2.1 Examine the Number of Distance Pairs and Maximum Lags between Residuals
The process of modeling spatial variability begins by obtaining the residuals from the original RCB analysis. Here PROC MIXED is used to fit the RCB model and output the model residuals to the SAS data set “residuals”.
ods html close;
proc mixed data=alliance;
class Rep Entry;
model Yield = Entry / outp=residuals;
random Rep;
run;
ods html;Examining and estimating spatial variability typically proceeds in several steps. In SAS, these can all be accomplished using PROC VARIOGRAM. In this first step, we summarize the potential distances (lags) between row/column positions. The nhclasses option sets a value for the number of lag classes or bins to try. Some trial and error here may be necessary in order to find a good setting, however, this value should be big enough to cover the range of possible distances between data row and column coordinates. Setting this initially to 40 was found to be a reasonable choice here. The novariogram option tells SAS to only look at these distances and not compute the empirical semivariance values yet.
/* Examine lag distance & maxlag */
ODS html GPATH = ".\img\" ;
proc variogram data=residuals;
compute novariogram nhclasses=40;
coordinates xc=row yc=col;
var resid;
run;
| Number of Observations Read | 224 |
|---|---|
| Number of Observations Used | 224 |
| Pairs Information | |
|---|---|
| Number of Lags | 41 |
| Lag Distance | 1.25 |
| Maximum Data Distance in row | 43.00 |
| Maximum Data Distance in col | 25.20 |
| Maximum Data Distance | 49.84 |
| Pairwise Distance Intervals | ||||
|---|---|---|---|---|
|
Lag Class |
Bounds | Number of Pairs |
Percentage of Pairs |
|
| 0 | 0.00 | 0.62 | 0 | 0.00% |
| 1 | 0.62 | 1.87 | 210 | 0.84% |
| 2 | 1.87 | 3.12 | 199 | 0.80% |
| 3 | 3.12 | 4.36 | 387 | 1.55% |
| 4 | 4.36 | 5.61 | 917 | 3.67% |
| 5 | 5.61 | 6.85 | 832 | 3.33% |
| 6 | 6.85 | 8.10 | 462 | 1.85% |
| 7 | 8.10 | 9.35 | 1571 | 6.29% |
| 8 | 9.35 | 10.59 | 1215 | 4.86% |
| 9 | 10.59 | 11.84 | 615 | 2.46% |
| 10 | 11.84 | 13.08 | 1252 | 5.01% |
| 11 | 13.08 | 14.33 | 1563 | 6.26% |
| 12 | 14.33 | 15.58 | 910 | 3.64% |
| 13 | 15.58 | 16.82 | 867 | 3.47% |
| 14 | 16.82 | 18.07 | 1805 | 7.23% |
| 15 | 18.07 | 19.31 | 1058 | 4.24% |
| 16 | 19.31 | 20.56 | 873 | 3.50% |
| 17 | 20.56 | 21.81 | 1352 | 5.41% |
| 18 | 21.81 | 23.05 | 935 | 3.74% |
| 19 | 23.05 | 24.30 | 943 | 3.78% |
| 20 | 24.30 | 25.54 | 517 | 2.07% |
| 21 | 25.54 | 26.79 | 1439 | 5.76% |
| 22 | 26.79 | 28.04 | 513 | 2.05% |
| 23 | 28.04 | 29.28 | 387 | 1.55% |
| 24 | 29.28 | 30.53 | 868 | 3.48% |
| 25 | 30.53 | 31.77 | 555 | 2.22% |
| 26 | 31.77 | 33.02 | 350 | 1.40% |
| 27 | 33.02 | 34.27 | 172 | 0.69% |
| 28 | 34.27 | 35.51 | 770 | 3.08% |
| 29 | 35.51 | 36.76 | 259 | 1.04% |
| 30 | 36.76 | 38.00 | 188 | 0.75% |
| 31 | 38.00 | 39.25 | 362 | 1.45% |
| 32 | 39.25 | 40.50 | 207 | 0.83% |
| 33 | 40.50 | 41.74 | 147 | 0.59% |
| 34 | 41.74 | 42.99 | 59 | 0.24% |
| 35 | 42.99 | 44.23 | 117 | 0.47% |
| 36 | 44.23 | 45.48 | 49 | 0.20% |
| 37 | 45.48 | 46.73 | 30 | 0.12% |
| 38 | 46.73 | 47.97 | 11 | 0.04% |
| 39 | 47.97 | 49.22 | 7 | 0.03% |
| 40 | 49.22 | 50.46 | 3 | 0.01% |
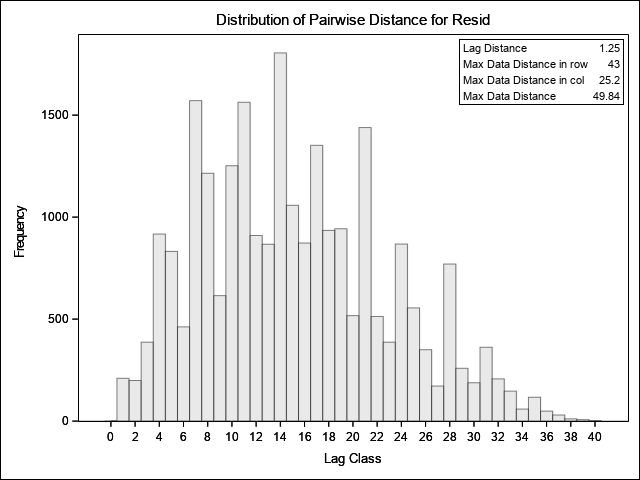
This output indicates that, with 40 bins, the minimum lag distance is approximately 1.2m and that the maximum lag distances vary from 25 to 43m in rows and columns, respectively. The histogram graphically displays the number of pairs at each lag distance bin. Ideally, we want bins that have at least 30 pairs to improve accuracy in estimation of the empirical semivariance. There is sufficient data here such that setting the maximum lag to 30 results in bins that have much more than 30 pairs and will provide an accurate estimate of semivariance.
6.2.2 Compute Moran’s I and Geary’s C.
Two common metrics of spatial correlation are Moran’s I and Geary’s c. Both these measures are essentially weighted correlations between pairs of observations and measure global and local variability, respectively. If no spatial correlation is present, Moran’s I has an expected value close to 0.0 while Geary’s C will be close to 1.0. The estimates for I and C can be computed and tested against these null values with the PROC VARIOGRAM code below.
ODS html GPATH = ".\img\" ;
proc variogram data=residuals plots(only)=moran ;
compute lagd=1.2 maxlag=30 novariogram autocorr(assum=nor) ;
coordinates xc=row yc=col;
var resid;
run;
| Number of Observations Read | 224 |
|---|---|
| Number of Observations Used | 224 |
| Pairs Information | |
|---|---|
| Number of Lags | 11 |
| Lag Distance | 4.98 |
| Maximum Data Distance in row | 43.00 |
| Maximum Data Distance in col | 25.20 |
| Maximum Data Distance | 49.84 |
| Pairwise Distance Intervals | ||||
|---|---|---|---|---|
|
Lag Class |
Bounds | Number of Pairs |
Percentage of Pairs |
|
| 0 | 0.00 | 2.49 | 409 | 1.64% |
| 1 | 2.49 | 7.48 | 2598 | 10.40% |
| 2 | 7.48 | 12.46 | 3757 | 15.04% |
| 3 | 12.46 | 17.44 | 5204 | 20.84% |
| 4 | 17.44 | 22.43 | 4738 | 18.97% |
| 5 | 22.43 | 27.41 | 3455 | 13.83% |
| 6 | 27.41 | 32.40 | 2241 | 8.97% |
| 7 | 32.40 | 37.38 | 1518 | 6.08% |
| 8 | 37.38 | 42.36 | 813 | 3.26% |
| 9 | 42.36 | 47.35 | 228 | 0.91% |
| 10 | 47.35 | 52.33 | 15 | 0.06% |
| Autocorrelation Statistics | ||||||
|---|---|---|---|---|---|---|
| Assumption | Coefficient | Observed | Expected | Std Dev | Z | Pr > |Z| |
| Normality | Moran's I | 0.330 | -0.00495 | 0.0836 | 4.01 | <.0001 |
| Normality | Geary's c | 0.614 | 1.00000 | 0.0901 | -4.29 | <.0001 |
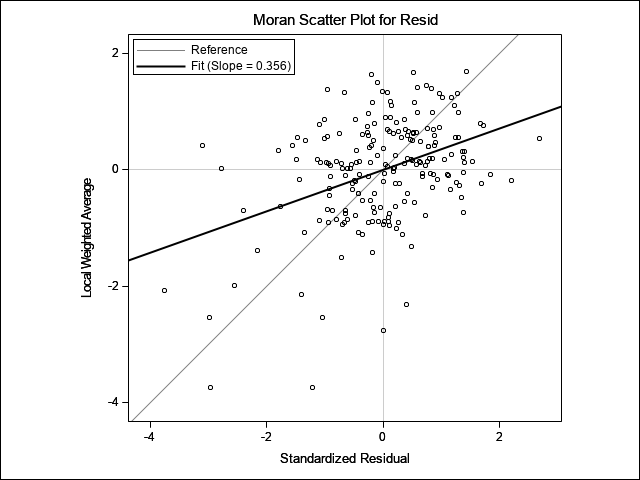
Both Moran’s I and Geary’s C are significant here, indicating the presence of spatial correlation. The accompanying scatter plot of residuals vs lag distance also show a positive correlation where residuals increase in magnitude with increasing distance between points.
6.3 Estimation and Modeling of Semivariance
6.3.1 Estimating Empirical Semivariance
Using the lag distance and maximum lag values obtained in the previous steps, we can now use the compute statement to estimate an empirical variogram as described in Section 2
ODS html GPATH = ".\img\" ;
proc variogram data=residuals plots(only)=(semivar);
coordinates xc=Col yc=Row;
compute lagd=1.2 maxlags=30;
var resid;
run;
| Number of Observations Read | 224 |
|---|---|
| Number of Observations Used | 224 |
| Empirical Semivariogram | |||
|---|---|---|---|
|
Lag Class |
Pair Count |
Average Distance |
Semivariance |
| 0 | 0 | . | . |
| 1 | 210 | 1.20 | 20.133 |
| 2 | 199 | 2.40 | 22.159 |
| 3 | 188 | 3.60 | 24.453 |
| 4 | 1116 | 4.64 | 21.737 |
| 5 | 832 | 6.01 | 25.005 |
| 6 | 462 | 7.32 | 28.096 |
| 7 | 1268 | 8.63 | 26.521 |
| 8 | 996 | 9.54 | 30.424 |
| 9 | 904 | 10.75 | 34.523 |
| 10 | 589 | 11.86 | 37.983 |
| 11 | 1812 | 13.14 | 32.064 |
| 12 | 1164 | 14.30 | 36.624 |
| 13 | 1021 | 15.67 | 41.839 |
| 14 | 1207 | 17.12 | 38.201 |
| 15 | 1329 | 17.97 | 38.456 |
| 16 | 988 | 19.19 | 41.319 |
| 17 | 691 | 20.46 | 47.746 |
| 18 | 1556 | 21.69 | 39.750 |
| 19 | 965 | 22.82 | 41.013 |
| 20 | 558 | 23.98 | 42.051 |
| 21 | 522 | 25.13 | 43.919 |
| 22 | 1273 | 26.21 | 35.351 |
| 23 | 588 | 27.55 | 42.812 |
| 24 | 312 | 28.69 | 43.350 |
| 25 | 868 | 30.13 | 37.531 |
| 26 | 555 | 31.11 | 42.186 |
| 27 | 350 | 32.41 | 48.901 |
| 28 | 172 | 33.60 | 56.251 |
| 29 | 704 | 34.68 | 33.781 |
| 30 | 325 | 35.91 | 45.543 |
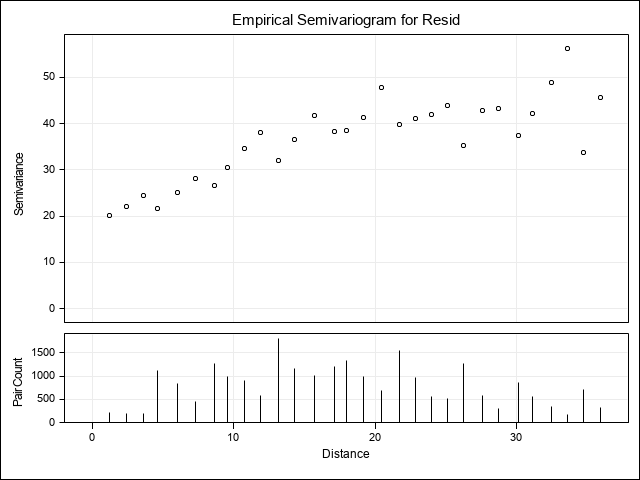
In the plot, the semi-variance increases as distance between points increases up to approximately 20m where it begins to level off. This type of pattern is very common for spatial relationships.
6.3.2 Fitting an Empirical Variogram Model
In Section 3, several theoretical variogram models were described. We can use PROC VARIOGRAM to fit and compare any number of these models. In the code below, the Gaussian, Exponential, Power, and Spherical models are fit using the model statement. By default when several models are listed, SAS will carry out a more sophisticated spatial modeling approach that uses combinations of these models. We, however, just want to focus on single model scenarios. Hence, the nest=1 option is given which restricts the variogram modeling to single models only.
ODS html GPATH = ".\img\" ;
proc variogram data=residuals plots(only)=(fitplot);
coordinates xc=Col yc=Row;
compute lagd=1.2 maxlags=30;
model form=auto(mlist=(gau, exp, pow, sph) nest=1);
var resid;
run;
| Number of Observations Read | 224 |
|---|---|
| Number of Observations Used | 224 |
| Empirical Semivariogram | |||
|---|---|---|---|
|
Lag Class |
Pair Count |
Average Distance |
Semivariance |
| 0 | 0 | . | . |
| 1 | 210 | 1.20 | 20.133 |
| 2 | 199 | 2.40 | 22.159 |
| 3 | 188 | 3.60 | 24.453 |
| 4 | 1116 | 4.64 | 21.737 |
| 5 | 832 | 6.01 | 25.005 |
| 6 | 462 | 7.32 | 28.096 |
| 7 | 1268 | 8.63 | 26.521 |
| 8 | 996 | 9.54 | 30.424 |
| 9 | 904 | 10.75 | 34.523 |
| 10 | 589 | 11.86 | 37.983 |
| 11 | 1812 | 13.14 | 32.064 |
| 12 | 1164 | 14.30 | 36.624 |
| 13 | 1021 | 15.67 | 41.839 |
| 14 | 1207 | 17.12 | 38.201 |
| 15 | 1329 | 17.97 | 38.456 |
| 16 | 988 | 19.19 | 41.319 |
| 17 | 691 | 20.46 | 47.746 |
| 18 | 1556 | 21.69 | 39.750 |
| 19 | 965 | 22.82 | 41.013 |
| 20 | 558 | 23.98 | 42.051 |
| 21 | 522 | 25.13 | 43.919 |
| 22 | 1273 | 26.21 | 35.351 |
| 23 | 588 | 27.55 | 42.812 |
| 24 | 312 | 28.69 | 43.350 |
| 25 | 868 | 30.13 | 37.531 |
| 26 | 555 | 31.11 | 42.186 |
| 27 | 350 | 32.41 | 48.901 |
| 28 | 172 | 33.60 | 56.251 |
| 29 | 704 | 34.68 | 33.781 |
| 30 | 325 | 35.91 | 45.543 |
| Semivariogram Model Fitting | |
|---|---|
| Model | Selection from 4 form combinations |
| Fit Summary | |||
|---|---|---|---|
| Class | Model |
Weighted SSE |
AIC |
| 1 | Gau | 90.90699 | 39.25919 |
| 2 | Sph | 95.41630 | 40.71156 |
| 3 | Exp | 111.51151 | 45.38791 |
| 4 | Pow | 131.80037 | 50.40273 |
|
Semivariogram Model Fitting |
|
|---|---|
| Name | Gaussian |
| Label | Gau |
| Model Information | |
|---|---|
| Parameter |
Initial Value |
| Nugget | 18.1070 |
| Scale | 27.0845 |
| Range | 17.9554 |
| Optimization Information | |
|---|---|
| Optimization Technique | Dual Quasi-Newton |
| Parameters in Optimization | 3 |
| Lower Boundaries | 3 |
| Upper Boundaries | 0 |
| Starting Values From | PROC |
| Parameter Estimates | |||||
|---|---|---|---|---|---|
| Parameter | Estimate |
Approx Std Error |
DF | t Value |
Approx Pr > |t| |
| Nugget | 19.6331 | 0.5918 | 27 | 33.18 | <.0001 |
| Scale | 22.0523 | 0.6610 | 27 | 33.36 | <.0001 |
| Range | 11.6661 | 0.4488 | 27 | 26.00 | <.0001 |
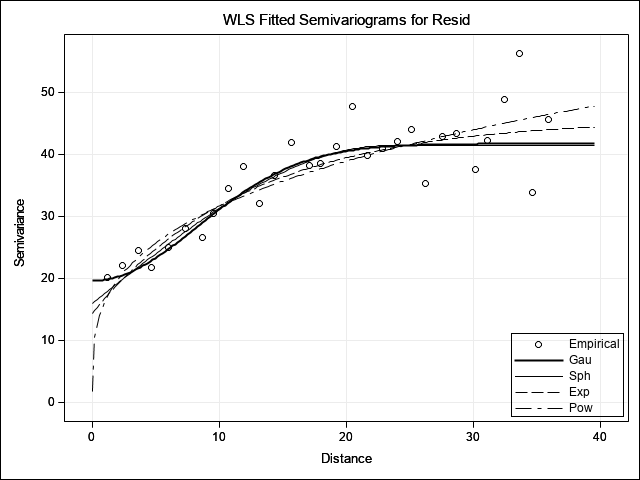
The table Fit Summary provides statistics on each variogram model fit. The AIC or SSE statistics can be used as a guide for model selection where smaller values indicate a better fit. For this data, the Gaussian model is rated as “best”, although the Spherical model is relatively similar. Given the numerically lower AIC and SSE values, however, PROC VARIOGRAM selects the Gaussian model and reports those semivariogram model parameter estimates in the final table. These parameter estimates will be used in the next step. The plot shown at the end provides a visual comparison each variogram model fit with the selected Gaussian model highlighted in bold.
6.4 Using the Estimated variogram in an Adjusted Analysis
The previous steps have:
1. Diagnosed the presence and pattern of spatial variability.
2. Identified and estimated the a model to describe the variability
The last step is to utilize this information in a statistical analysis as outlined at the beginning of Section 3. To do this, the linear mixed model procedure PROC MIXED will be used to incorporate the spatial variability into the linear model variance-covariance matrix, assuming the variogram model identified above.
As an initial step, for comparison purposes, the unadjusted RCB model is run first and the means saved in data set “NIN_RCBD_means”.
6.4.1 Unadjusted RCBD Model
/** Fit unadjusted RCBD model **/
ODS html GPATH = ".\img\" ;
proc mixed data=alliance ;
class entry rep;
model yield = entry ;
random rep;
lsmeans entry/cl;
ods output LSMeans=NIN_RCBD_means;
title1 'NIN data: RCBD';
run;| NIN data: RCBD |
| Model Information | |
|---|---|
| Data Set | WORK.ALLIANCE |
| Dependent Variable | yield |
| Covariance Structure | Variance Components |
| Estimation Method | REML |
| Residual Variance Method | Profile |
| Fixed Effects SE Method | Model-Based |
| Degrees of Freedom Method | Containment |
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| entry | 56 | Arapahoe Brule Buckskin Centura Centurk7 Cheyenne Cody Colt Gage Homestea KS831374 Lancer Lancota NE83404 NE83406 NE83407 NE83432 NE83498 NE83T12 NE84557 NE85556 NE85623 NE86482 NE86501 NE86503 NE86507 NE86509 NE86527 NE86582 NE86606 NE86607 NE86T666 NE87403 NE87408 NE87409 NE87446 NE87451 NE87457 NE87463 NE87499 NE87512 NE87513 NE87522 NE87612 NE87613 NE87615 NE87619 NE87627 Norkan Redland Roughrid Scout66 Siouxlan TAM107 TAM200 Vona |
| rep | 4 | R1 R2 R3 R4 |
| Dimensions | |
|---|---|
| Covariance Parameters | 2 |
| Columns in X | 57 |
| Columns in Z | 4 |
| Subjects | 1 |
| Max Obs per Subject | 224 |
| Number of Observations | |
|---|---|
| Number of Observations Read | 224 |
| Number of Observations Used | 224 |
| Number of Observations Not Used | 0 |
| Iteration History | |||
|---|---|---|---|
| Iteration | Evaluations | -2 Res Log Like | Criterion |
| 0 | 1 | 1240.74178756 | |
| 1 | 1 | 1217.70153249 | 0.00000000 |
| Convergence criteria met. |
| Covariance Parameter Estimates | |
|---|---|
| Cov Parm | Estimate |
| rep | 9.8829 |
| Residual | 49.5824 |
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | 1217.7 |
| AIC (Smaller is Better) | 1221.7 |
| AICC (Smaller is Better) | 1221.8 |
| BIC (Smaller is Better) | 1220.5 |
| Type 3 Tests of Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| entry | 55 | 165 | 0.88 | 0.7119 |
| Least Squares Means | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Effect | entry | Estimate |
Standard Error |
DF | t Value | Pr > |t| | Alpha | Lower | Upper |
| entry | Arapahoe | 29.4375 | 3.8557 | 165 | 7.63 | <.0001 | 0.05 | 21.8247 | 37.0503 |
| entry | Brule | 26.0750 | 3.8557 | 165 | 6.76 | <.0001 | 0.05 | 18.4622 | 33.6878 |
| entry | Buckskin | 25.5625 | 3.8557 | 165 | 6.63 | <.0001 | 0.05 | 17.9497 | 33.1753 |
| entry | Centura | 21.6500 | 3.8557 | 165 | 5.62 | <.0001 | 0.05 | 14.0372 | 29.2628 |
| entry | Centurk7 | 30.3000 | 3.8557 | 165 | 7.86 | <.0001 | 0.05 | 22.6872 | 37.9128 |
| entry | Cheyenne | 28.0625 | 3.8557 | 165 | 7.28 | <.0001 | 0.05 | 20.4497 | 35.6753 |
| entry | Cody | 21.2125 | 3.8557 | 165 | 5.50 | <.0001 | 0.05 | 13.5997 | 28.8253 |
| entry | Colt | 27.0000 | 3.8557 | 165 | 7.00 | <.0001 | 0.05 | 19.3872 | 34.6128 |
| entry | Gage | 24.5125 | 3.8557 | 165 | 6.36 | <.0001 | 0.05 | 16.8997 | 32.1253 |
| entry | Homestea | 27.6375 | 3.8557 | 165 | 7.17 | <.0001 | 0.05 | 20.0247 | 35.2503 |
| entry | KS831374 | 24.1250 | 3.8557 | 165 | 6.26 | <.0001 | 0.05 | 16.5122 | 31.7378 |
| entry | Lancer | 28.5625 | 3.8557 | 165 | 7.41 | <.0001 | 0.05 | 20.9497 | 36.1753 |
| entry | Lancota | 26.5500 | 3.8557 | 165 | 6.89 | <.0001 | 0.05 | 18.9372 | 34.1628 |
| entry | NE83404 | 27.3875 | 3.8557 | 165 | 7.10 | <.0001 | 0.05 | 19.7747 | 35.0003 |
| entry | NE83406 | 24.2750 | 3.8557 | 165 | 6.30 | <.0001 | 0.05 | 16.6622 | 31.8878 |
| entry | NE83407 | 22.6875 | 3.8557 | 165 | 5.88 | <.0001 | 0.05 | 15.0747 | 30.3003 |
| entry | NE83432 | 19.7250 | 3.8557 | 165 | 5.12 | <.0001 | 0.05 | 12.1122 | 27.3378 |
| entry | NE83498 | 30.1250 | 3.8557 | 165 | 7.81 | <.0001 | 0.05 | 22.5122 | 37.7378 |
| entry | NE83T12 | 21.5625 | 3.8557 | 165 | 5.59 | <.0001 | 0.05 | 13.9497 | 29.1753 |
| entry | NE84557 | 20.5250 | 3.8557 | 165 | 5.32 | <.0001 | 0.05 | 12.9122 | 28.1378 |
| entry | NE85556 | 26.3875 | 3.8557 | 165 | 6.84 | <.0001 | 0.05 | 18.7747 | 34.0003 |
| entry | NE85623 | 21.7250 | 3.8557 | 165 | 5.63 | <.0001 | 0.05 | 14.1122 | 29.3378 |
| entry | NE86482 | 24.2875 | 3.8557 | 165 | 6.30 | <.0001 | 0.05 | 16.6747 | 31.9003 |
| entry | NE86501 | 30.9375 | 3.8557 | 165 | 8.02 | <.0001 | 0.05 | 23.3247 | 38.5503 |
| entry | NE86503 | 32.6500 | 3.8557 | 165 | 8.47 | <.0001 | 0.05 | 25.0372 | 40.2628 |
| entry | NE86507 | 23.7875 | 3.8557 | 165 | 6.17 | <.0001 | 0.05 | 16.1747 | 31.4003 |
| entry | NE86509 | 26.8500 | 3.8557 | 165 | 6.96 | <.0001 | 0.05 | 19.2372 | 34.4628 |
| entry | NE86527 | 22.0125 | 3.8557 | 165 | 5.71 | <.0001 | 0.05 | 14.3997 | 29.6253 |
| entry | NE86582 | 24.5375 | 3.8557 | 165 | 6.36 | <.0001 | 0.05 | 16.9247 | 32.1503 |
| entry | NE86606 | 29.7625 | 3.8557 | 165 | 7.72 | <.0001 | 0.05 | 22.1497 | 37.3753 |
| entry | NE86607 | 29.3250 | 3.8557 | 165 | 7.61 | <.0001 | 0.05 | 21.7122 | 36.9378 |
| entry | NE86T666 | 21.5375 | 3.8557 | 165 | 5.59 | <.0001 | 0.05 | 13.9247 | 29.1503 |
| entry | NE87403 | 25.1250 | 3.8557 | 165 | 6.52 | <.0001 | 0.05 | 17.5122 | 32.7378 |
| entry | NE87408 | 26.3000 | 3.8557 | 165 | 6.82 | <.0001 | 0.05 | 18.6872 | 33.9128 |
| entry | NE87409 | 21.3750 | 3.8557 | 165 | 5.54 | <.0001 | 0.05 | 13.7622 | 28.9878 |
| entry | NE87446 | 27.6750 | 3.8557 | 165 | 7.18 | <.0001 | 0.05 | 20.0622 | 35.2878 |
| entry | NE87451 | 24.6125 | 3.8557 | 165 | 6.38 | <.0001 | 0.05 | 16.9997 | 32.2253 |
| entry | NE87457 | 23.9125 | 3.8557 | 165 | 6.20 | <.0001 | 0.05 | 16.2997 | 31.5253 |
| entry | NE87463 | 25.9125 | 3.8557 | 165 | 6.72 | <.0001 | 0.05 | 18.2997 | 33.5253 |
| entry | NE87499 | 20.4125 | 3.8557 | 165 | 5.29 | <.0001 | 0.05 | 12.7997 | 28.0253 |
| entry | NE87512 | 23.2500 | 3.8557 | 165 | 6.03 | <.0001 | 0.05 | 15.6372 | 30.8628 |
| entry | NE87513 | 26.8125 | 3.8557 | 165 | 6.95 | <.0001 | 0.05 | 19.1997 | 34.4253 |
| entry | NE87522 | 25.0000 | 3.8557 | 165 | 6.48 | <.0001 | 0.05 | 17.3872 | 32.6128 |
| entry | NE87612 | 21.8000 | 3.8557 | 165 | 5.65 | <.0001 | 0.05 | 14.1872 | 29.4128 |
| entry | NE87613 | 29.4000 | 3.8557 | 165 | 7.63 | <.0001 | 0.05 | 21.7872 | 37.0128 |
| entry | NE87615 | 25.6875 | 3.8557 | 165 | 6.66 | <.0001 | 0.05 | 18.0747 | 33.3003 |
| entry | NE87619 | 31.2625 | 3.8557 | 165 | 8.11 | <.0001 | 0.05 | 23.6497 | 38.8753 |
| entry | NE87627 | 23.2250 | 3.8557 | 165 | 6.02 | <.0001 | 0.05 | 15.6122 | 30.8378 |
| entry | Norkan | 24.4125 | 3.8557 | 165 | 6.33 | <.0001 | 0.05 | 16.7997 | 32.0253 |
| entry | Redland | 30.5000 | 3.8557 | 165 | 7.91 | <.0001 | 0.05 | 22.8872 | 38.1128 |
| entry | Roughrid | 21.1875 | 3.8557 | 165 | 5.50 | <.0001 | 0.05 | 13.5747 | 28.8003 |
| entry | Scout66 | 27.5250 | 3.8557 | 165 | 7.14 | <.0001 | 0.05 | 19.9122 | 35.1378 |
| entry | Siouxlan | 30.1125 | 3.8557 | 165 | 7.81 | <.0001 | 0.05 | 22.4997 | 37.7253 |
| entry | TAM107 | 28.4000 | 3.8557 | 165 | 7.37 | <.0001 | 0.05 | 20.7872 | 36.0128 |
| entry | TAM200 | 21.2375 | 3.8557 | 165 | 5.51 | <.0001 | 0.05 | 13.6247 | 28.8503 |
| entry | Vona | 23.6000 | 3.8557 | 165 | 6.12 | <.0001 | 0.05 | 15.9872 | 31.2128 |
The F-statistic and accompanying large p-value for ‘gen’ indicate no differences between cultivars. This is surprising for a variety trial. The fit statistics (e.g. AIC) are helpful for model comparison, although on their own, these metrics do not have much meaning. The AIC value for this standard RCB analysis is 1221.7; let’s compare that number to that for the spatially-adjusted models (in the case of AIC, smaller numbers indicate a better fitting model). Another point of comparison is the standard error of the means: 3.8557.
The next step is to try a spatially-adjusted model. This is done by adding a repeated statement specifying the spatial model form (Gaussian, type=sp(gau)) and the variables related to spatial positions in the study (row, col). The local option tells SAS to use a nugget parameter in the Gaussian variogram model. The second additional statement is parms. This gives SAS a starting place for estimating the range, sill, and nugget parameters. The values given here are taken from the PROC VARIOGRAM output above. While this step can be omitted, it is recommended not to do so because, on its own, PROC MIXED can often have trouble narrowing in on reasonable parameter estimates. One reason for the preceding variogram steps was to obtain these parameter values so PROC MIXED has a good starting place for estimation. As before, the estimated means (adjusted now) are saved, this time in data set “NIN_Spatial_means”.
6.4.2 RCB Model with Spatial Covariance
/** Fit Gaussian adjusted model **/
/** Parms statement order: Range, Sill, Nugget **/
/** Model option "local" forces nugget into model **/
ODS html GPATH = ".\img\" ;
proc mixed data=alliance maxiter=150;
class entry;
model yield = entry /ddfm=kr;
repeated/subject=intercept type=sp(gau) (Row Col) local;
parms (11) (22) (19);
lsmeans entry/cl;
ods output LSMeans=NIN_Spatial_means;
title1 'NIN data: Gaussian Spatial Adjustment';
run;
| NIN data: Gaussian Spatial Adjustment |
| Model Information | |
|---|---|
| Data Set | WORK.ALLIANCE |
| Dependent Variable | yield |
| Covariance Structure | Spatial Gaussian |
| Subject Effect | Intercept |
| Estimation Method | REML |
| Residual Variance Method | Profile |
| Fixed Effects SE Method | Kenward-Roger |
| Degrees of Freedom Method | Kenward-Roger |
| Class Level Information | ||
|---|---|---|
| Class | Levels | Values |
| entry | 56 | Arapahoe Brule Buckskin Centura Centurk7 Cheyenne Cody Colt Gage Homestea KS831374 Lancer Lancota NE83404 NE83406 NE83407 NE83432 NE83498 NE83T12 NE84557 NE85556 NE85623 NE86482 NE86501 NE86503 NE86507 NE86509 NE86527 NE86582 NE86606 NE86607 NE86T666 NE87403 NE87408 NE87409 NE87446 NE87451 NE87457 NE87463 NE87499 NE87512 NE87513 NE87522 NE87612 NE87613 NE87615 NE87619 NE87627 Norkan Redland Roughrid Scout66 Siouxlan TAM107 TAM200 Vona |
| Dimensions | |
|---|---|
| Covariance Parameters | 3 |
| Columns in X | 57 |
| Columns in Z | 0 |
| Subjects | 1 |
| Max Obs per Subject | 224 |
| Number of Observations | |
|---|---|
| Number of Observations Read | 224 |
| Number of Observations Used | 224 |
| Number of Observations Not Used | 0 |
| Parameter Search | |||||
|---|---|---|---|---|---|
| CovP1 | CovP2 | CovP3 | Variance | Res Log Like | -2 Res Log Like |
| 11.0000 | 22.0000 | 19.0000 | 25.3339 | -555.0999 | 1110.1998 |
| Iteration History | |||
|---|---|---|---|
| Iteration | Evaluations | -2 Res Log Like | Criterion |
| 1 | 2 | 1098.19938899 | 0.55691103 |
| 2 | 2 | 1073.55589724 | 0.01698755 |
| 3 | 2 | 1070.40479989 | 0.00621291 |
| 4 | 1 | 1067.58247826 | 0.00103523 |
| 5 | 1 | 1067.13250829 | 0.00007623 |
| 6 | 1 | 1067.10196425 | 0.00000066 |
| 7 | 1 | 1067.10171375 | 0.00000000 |
| Convergence criteria met. |
| Covariance Parameter Estimates | ||
|---|---|---|
| Cov Parm | Subject | Estimate |
| Variance | Intercept | 43.3874 |
| SP(GAU) | Intercept | 10.7004 |
| Residual | 15.3580 | |
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | 1067.1 |
| AIC (Smaller is Better) | 1073.1 |
| AICC (Smaller is Better) | 1073.2 |
| BIC (Smaller is Better) | 1082.5 |
| PARMS Model Likelihood Ratio Test | ||
|---|---|---|
| DF | Chi-Square | Pr > ChiSq |
| 2 | 43.10 | <.0001 |
| Type 3 Tests of Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Num DF | Den DF | F Value | Pr > F |
| entry | 55 | 136 | 1.84 | 0.0024 |
| Least Squares Means | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Effect | entry | Estimate |
Standard Error |
DF | t Value | Pr > |t| | Alpha | Lower | Upper |
| entry | Arapahoe | 27.0657 | 3.2760 | 18.5 | 8.26 | <.0001 | 0.05 | 20.1964 | 33.9351 |
| entry | Brule | 27.0363 | 3.2912 | 18.3 | 8.21 | <.0001 | 0.05 | 20.1295 | 33.9430 |
| entry | Buckskin | 35.6362 | 3.3341 | 19.5 | 10.69 | <.0001 | 0.05 | 28.6692 | 42.6032 |
| entry | Centura | 25.4196 | 3.2861 | 17.9 | 7.74 | <.0001 | 0.05 | 18.5131 | 32.3261 |
| entry | Centurk7 | 27.2432 | 3.2915 | 18.2 | 8.28 | <.0001 | 0.05 | 20.3345 | 34.1519 |
| entry | Cheyenne | 25.0592 | 3.2843 | 18.3 | 7.63 | <.0001 | 0.05 | 18.1660 | 31.9524 |
| entry | Cody | 24.0971 | 3.2733 | 18.5 | 7.36 | <.0001 | 0.05 | 17.2346 | 30.9596 |
| entry | Colt | 25.7772 | 3.3473 | 18.5 | 7.70 | <.0001 | 0.05 | 18.7570 | 32.7974 |
| entry | Gage | 25.0110 | 3.2920 | 18 | 7.60 | <.0001 | 0.05 | 18.0941 | 31.9279 |
| entry | Homestea | 22.1425 | 3.2711 | 19.1 | 6.77 | <.0001 | 0.05 | 15.2994 | 28.9856 |
| entry | KS831374 | 26.9109 | 3.2537 | 18.8 | 8.27 | <.0001 | 0.05 | 20.0954 | 33.7263 |
| entry | Lancer | 23.6478 | 3.2891 | 18.2 | 7.19 | <.0001 | 0.05 | 16.7428 | 30.5528 |
| entry | Lancota | 21.7590 | 3.3014 | 18.5 | 6.59 | <.0001 | 0.05 | 14.8367 | 28.6814 |
| entry | NE83404 | 25.4502 | 3.2997 | 18.4 | 7.71 | <.0001 | 0.05 | 18.5282 | 32.3721 |
| entry | NE83406 | 26.5136 | 3.2808 | 18.5 | 8.08 | <.0001 | 0.05 | 19.6355 | 33.3918 |
| entry | NE83407 | 26.0105 | 3.3019 | 19.7 | 7.88 | <.0001 | 0.05 | 19.1168 | 32.9041 |
| entry | NE83432 | 22.3448 | 3.2852 | 18.3 | 6.80 | <.0001 | 0.05 | 15.4503 | 29.2394 |
| entry | NE83498 | 29.7725 | 3.3042 | 18.3 | 9.01 | <.0001 | 0.05 | 22.8396 | 36.7055 |
| entry | NE83T12 | 21.9692 | 3.3222 | 18.3 | 6.61 | <.0001 | 0.05 | 14.9968 | 28.9416 |
| entry | NE84557 | 21.8553 | 3.2802 | 18.4 | 6.66 | <.0001 | 0.05 | 14.9758 | 28.7347 |
| entry | NE85556 | 28.6144 | 3.2887 | 18.1 | 8.70 | <.0001 | 0.05 | 21.7073 | 35.5216 |
| entry | NE85623 | 24.2722 | 3.2740 | 18.4 | 7.41 | <.0001 | 0.05 | 17.4046 | 31.1398 |
| entry | NE86482 | 24.7351 | 3.2632 | 18.7 | 7.58 | <.0001 | 0.05 | 17.8977 | 31.5725 |
| entry | NE86501 | 25.4990 | 3.2964 | 18.6 | 7.74 | <.0001 | 0.05 | 18.5891 | 32.4090 |
| entry | NE86503 | 27.8735 | 3.3194 | 18.1 | 8.40 | <.0001 | 0.05 | 20.9011 | 34.8459 |
| entry | NE86507 | 27.8365 | 3.3336 | 17.9 | 8.35 | <.0001 | 0.05 | 20.8308 | 34.8421 |
| entry | NE86509 | 22.7981 | 3.3010 | 18.4 | 6.91 | <.0001 | 0.05 | 15.8736 | 29.7226 |
| entry | NE86527 | 25.4641 | 3.3056 | 18 | 7.70 | <.0001 | 0.05 | 18.5193 | 32.4089 |
| entry | NE86582 | 24.0511 | 3.3056 | 18 | 7.28 | <.0001 | 0.05 | 17.1053 | 30.9969 |
| entry | NE86606 | 28.0865 | 3.3221 | 17.9 | 8.45 | <.0001 | 0.05 | 21.1042 | 35.0688 |
| entry | NE86607 | 26.4158 | 3.3368 | 18.2 | 7.92 | <.0001 | 0.05 | 19.4105 | 33.4210 |
| entry | NE86T666 | 17.8942 | 3.2827 | 18.1 | 5.45 | <.0001 | 0.05 | 10.9996 | 24.7887 |
| entry | NE87403 | 22.3379 | 3.2971 | 19 | 6.77 | <.0001 | 0.05 | 15.4380 | 29.2378 |
| entry | NE87408 | 25.1692 | 3.3024 | 18.8 | 7.62 | <.0001 | 0.05 | 18.2524 | 32.0860 |
| entry | NE87409 | 26.8337 | 3.3081 | 18.4 | 8.11 | <.0001 | 0.05 | 19.8953 | 33.7722 |
| entry | NE87446 | 22.6374 | 3.2889 | 17.9 | 6.88 | <.0001 | 0.05 | 15.7249 | 29.5500 |
| entry | NE87451 | 24.8679 | 3.3107 | 18.4 | 7.51 | <.0001 | 0.05 | 17.9220 | 31.8138 |
| entry | NE87457 | 24.1953 | 3.3139 | 17.8 | 7.30 | <.0001 | 0.05 | 17.2285 | 31.1622 |
| entry | NE87463 | 24.0642 | 3.2973 | 18.1 | 7.30 | <.0001 | 0.05 | 17.1393 | 30.9892 |
| entry | NE87499 | 22.4493 | 3.2968 | 17.6 | 6.81 | <.0001 | 0.05 | 15.5131 | 29.3855 |
| entry | NE87512 | 23.2063 | 3.2933 | 18.5 | 7.05 | <.0001 | 0.05 | 16.3019 | 30.1108 |
| entry | NE87513 | 22.1161 | 3.2875 | 18.1 | 6.73 | <.0001 | 0.05 | 15.2123 | 29.0200 |
| entry | NE87522 | 20.3275 | 3.2839 | 18.1 | 6.19 | <.0001 | 0.05 | 13.4302 | 27.2248 |
| entry | NE87612 | 28.9563 | 3.2808 | 18.5 | 8.83 | <.0001 | 0.05 | 22.0772 | 35.8354 |
| entry | NE87613 | 28.2664 | 3.3038 | 18.2 | 8.56 | <.0001 | 0.05 | 21.3296 | 35.2031 |
| entry | NE87615 | 24.6426 | 3.2876 | 18 | 7.50 | <.0001 | 0.05 | 17.7343 | 31.5509 |
| entry | NE87619 | 29.3455 | 3.2904 | 18.1 | 8.92 | <.0001 | 0.05 | 22.4359 | 36.2550 |
| entry | NE87627 | 19.7546 | 3.3239 | 18.2 | 5.94 | <.0001 | 0.05 | 12.7760 | 26.7333 |
| entry | Norkan | 23.2929 | 3.2853 | 18 | 7.09 | <.0001 | 0.05 | 16.3915 | 30.1942 |
| entry | Redland | 28.2315 | 3.2653 | 18.7 | 8.65 | <.0001 | 0.05 | 21.3899 | 35.0732 |
| entry | Roughrid | 26.3372 | 3.3674 | 18.3 | 7.82 | <.0001 | 0.05 | 19.2718 | 33.4025 |
| entry | Scout66 | 26.6969 | 3.2851 | 18.3 | 8.13 | <.0001 | 0.05 | 19.8032 | 33.5906 |
| entry | Siouxlan | 26.4493 | 3.3019 | 18.1 | 8.01 | <.0001 | 0.05 | 19.5163 | 33.3823 |
| entry | TAM107 | 23.1066 | 3.2840 | 18.5 | 7.04 | <.0001 | 0.05 | 16.2203 | 29.9930 |
| entry | TAM200 | 19.2002 | 3.2840 | 18.1 | 5.85 | <.0001 | 0.05 | 12.3046 | 26.0959 |
| entry | Vona | 25.6633 | 3.3045 | 18.2 | 7.77 | <.0001 | 0.05 | 18.7257 | 32.6009 |
From this output, we see the AIC value is now 1073.1. This is substantially lower than the AIC=1221.7 from the unadjusted model indicating a better model fit. Additionally, the gen effect in the model now has a low p-value (p=0.0024) giving evidence for differences among cultivars where we did not see them previously. Lastly, the standard errors for the means are now lower than those of the unadjusted model indicating an increased precision in mean estimation.
6.4.3 Other Spatial Adjustments
6.4.3.1 RCB Model with Row and Column Covariates
Spatial variability might also be addressed by including covariates in the model for row and/or column trends. The following script fits this model where rows and columns enter the model as continuous effects (not in the class statement).
/** Row column model **/
ods html close;
proc mixed data=alliance ;
class entry rep;
model yield = entry row col/ddfm=kr;
random rep;
lsmeans entry/cl;
ods output LSMeans=NIN_row_col_means;
title1 'NIN data: RCBD';
run;
ods html;
6.4.3.2 RCB with Spline Adjustment
Polynomial splines are an additional method for spatial adjustment and represent a more non-parametric method that does not rely on estimation or modeling of variograms. Instead, it uses the raw data and residuals to fit a surface to the spatial data and adjust the variance covariance matrix accordingly. PROC MIXED, however, does not have an option for this, so the code below moves to PROC GLIMMIX, the generalized Linear Mixed Model procedure in SAS. The syntax for the two procedures are very similar, so, in this example adapted from (Stroup 2013), only a few changes are required. First, a random statement is added for rows and columns with a “type=rsmooth” option. This fits a spline to the residuals over the row and column axes of the study area. Second, an “effect” statement is given defining a new covariate, sm_r, which is a splined surface for raw yields over the study area. When entered into the model as a continuous covariate, this centers the data such that the resulting model residuals will have a mean of zero.
ods html close;
/** Fit RCBD model with Spline **/
proc glimmix data=alliance ;
class entry rep;
effect sp_r = spline(row col);
model yield = entry sp_r/ddfm=kr;
random row col/type=rsmooth;
lsmeans entry/cl;
ods output LSMeans=NIN_smooth_means;
title1 'NIN data: RCBD';
run;
ods html;
6.5 Compare Estimated Means
Presence of spatial variability can also bias the mean estimates. In this data, this would result in the means ranking incorrectly relative to one another under the unadjusted model. The steps below combine the saved means from the unadjusted and spatially adjusted models above so that we can see how the ranking changes.
data NIN_RCBD_means (drop=tvalue probt alpha estimate stderr lower upper df);
set NIN_RCBD_means;
RCB_est = estimate;
RCB_se = stderr;
run;
data NIN_Spatial_means (drop=tvalue probt alpha estimate stderr lower upper df);
set NIN_Spatial_means;
Sp_est = estimate;
Sp_se = stderr;
run;
proc sort data=NIN_RCBD_means;
by entry;
run;
proc sort data=NIN_Spatial_means;
by entry;
run;
data compare;
merge NIN_RCBD_means NIN_Spatial_means;
by entry;
run;
proc rank data=compare out=compare descending;
var RCB_est Sp_est;
ranks RCB_Rank Sp_Rank;
run;
proc sort data=compare;
by Sp_rank;
run;
proc print data=compare(obs=15);
var entry rcb_est Sp_est rcb_se sp_se rcb_rank sp_rank;
run;| Obs | entry | RCB_est | Sp_est | RCB_se | Sp_se | RCB_Rank | Sp_Rank |
|---|---|---|---|---|---|---|---|
| 1 | Buckskin | 25.5625 | 35.6362 | 3.85569 | 3.33413 | 28 | 1 |
| 2 | NE83498 | 30.1250 | 29.7725 | 3.85569 | 3.30417 | 6 | 2 |
| 3 | NE87619 | 31.2625 | 29.3455 | 3.85569 | 3.29042 | 2 | 3 |
| 4 | NE87612 | 21.8000 | 28.9563 | 3.85569 | 3.28084 | 45 | 4 |
| 5 | NE85556 | 26.3875 | 28.6144 | 3.85569 | 3.28870 | 23 | 5 |
| 6 | NE87613 | 29.4000 | 28.2664 | 3.85569 | 3.30377 | 10 | 6 |
| 7 | Redland | 30.5000 | 28.2315 | 3.85569 | 3.26531 | 4 | 7 |
| 8 | NE86606 | 29.7625 | 28.0865 | 3.85569 | 3.32212 | 8 | 8 |
| 9 | NE86503 | 32.6500 | 27.8735 | 3.85569 | 3.31941 | 1 | 9 |
| 10 | NE86507 | 23.7875 | 27.8365 | 3.85569 | 3.33361 | 39 | 10 |
| 11 | Centurk7 | 30.3000 | 27.2432 | 3.85569 | 3.29145 | 5 | 11 |
| 12 | Arapahoe | 29.4375 | 27.0657 | 3.85569 | 3.27598 | 9 | 12 |
| 13 | Brule | 26.0750 | 27.0363 | 3.85569 | 3.29121 | 25 | 13 |
| 14 | KS831374 | 24.1250 | 26.9109 | 3.85569 | 3.25366 | 37 | 14 |
| 15 | NE87409 | 21.3750 | 26.8337 | 3.85569 | 3.30810 | 50 | 15 |
This comparison of the first 15 means shows some clear changes in rank. In his discussion of this data, Stroup (Stroup 2013), writes:
… Buckskin was a variety known to be a high-yielding benchmark; its mediocre mean yield in the RCB analysis despite being observed in the field outperforming all varieties in the vicinity was one symptom that the RCB analysis was giving nonsense results.
— Stroup (2013)
This high expectation for Buckskin is realized in the adjusted analysis. Likewise, other varieties, such as NE87612 that ranked near the bottom in the RCB analysis, ranked as number 4 in the adjusted analysis. Of the top 10 varieties identified in the adjusted analysis, only 6 are found in the top 10 of the unadjusted RCB analysis. These changes in rank illustrate why consideration of spatial variability in agricultural trials is important.